Download
Subject 1
Subject 2
Subject 3
Subject 4
Pose Preview
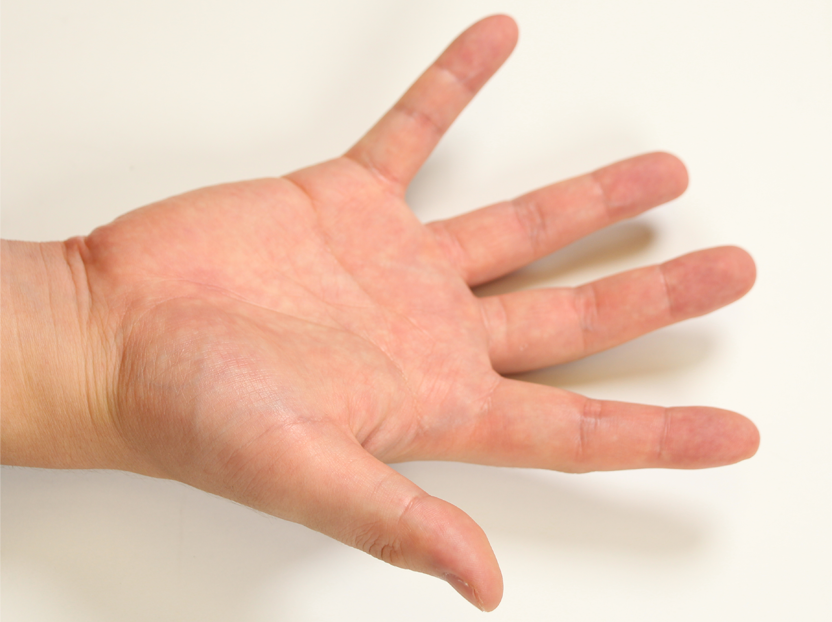 Pose 01 |  Pose 02 | 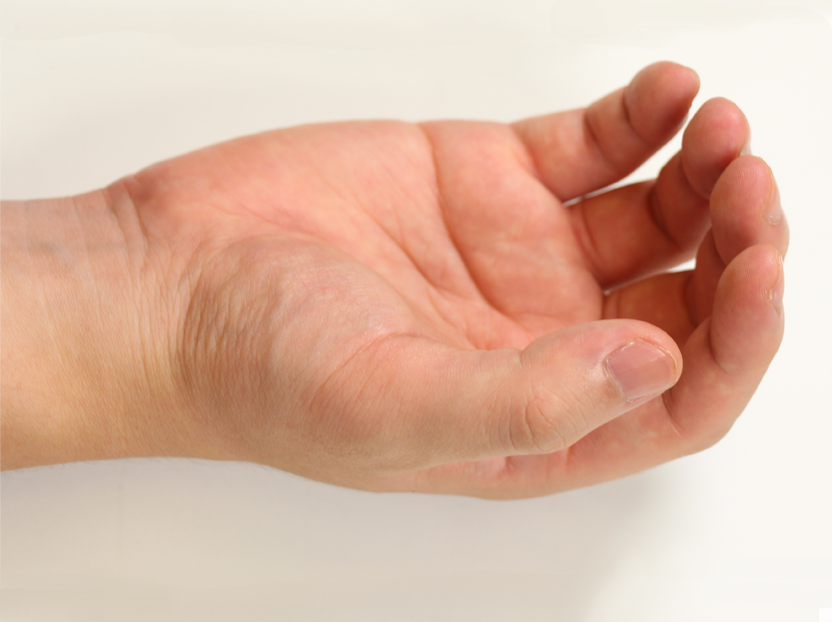 Pose 03 | 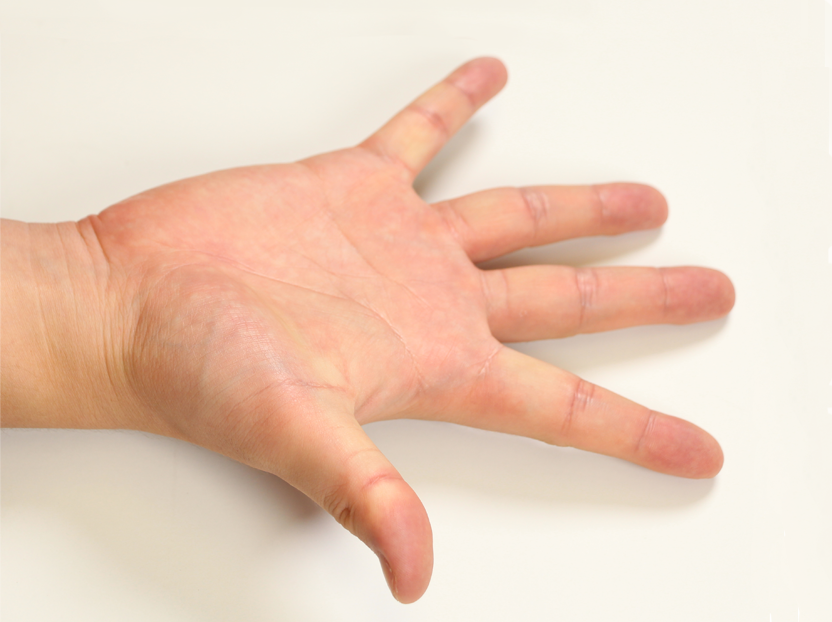 Pose 04 | 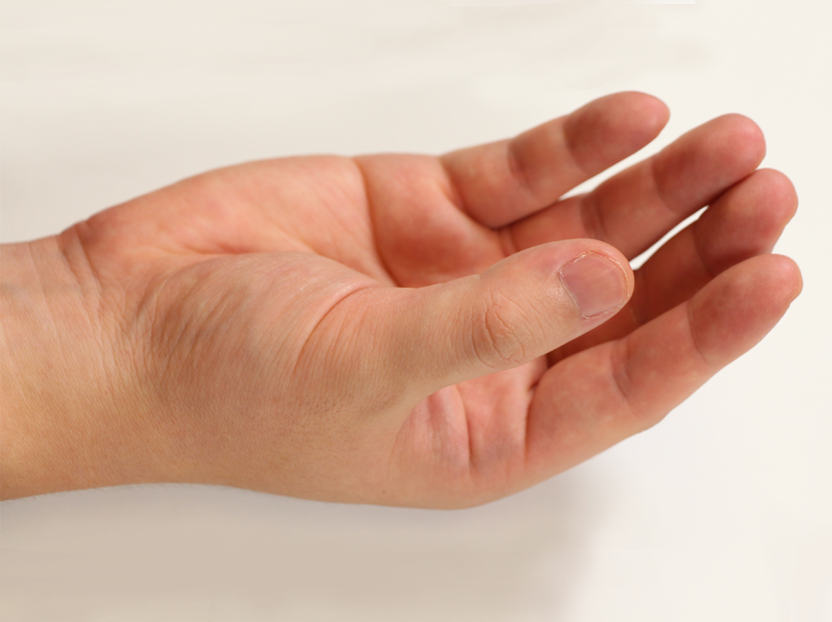 Pose 05 | 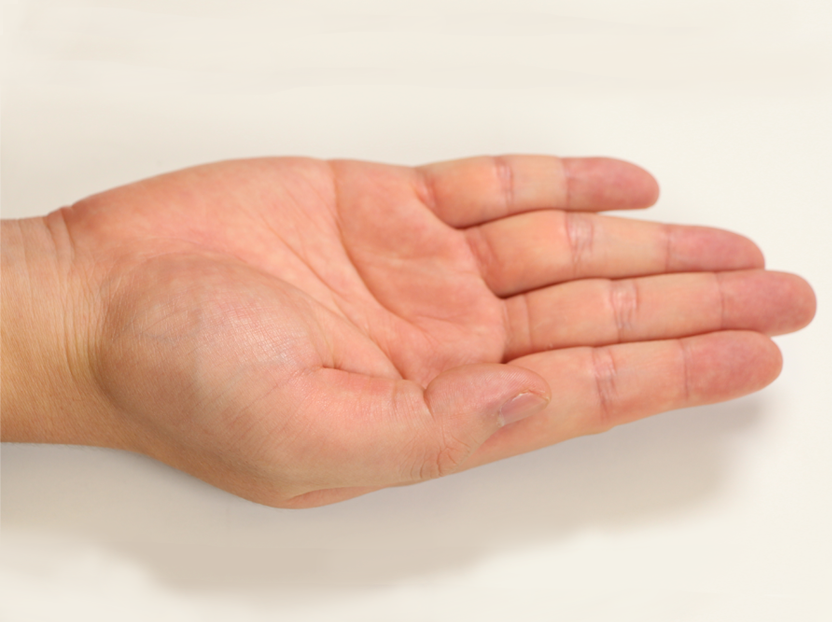 Pose 06 |
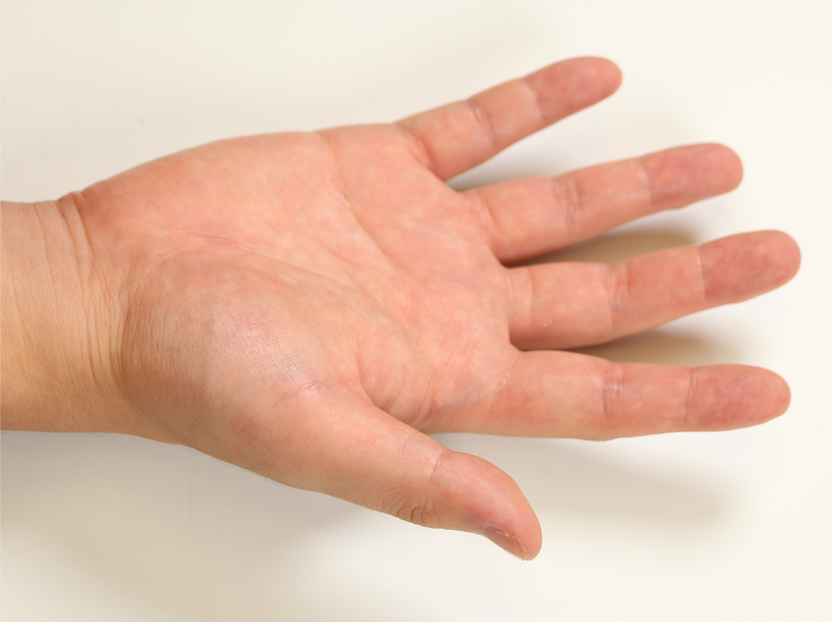 Pose 07 | 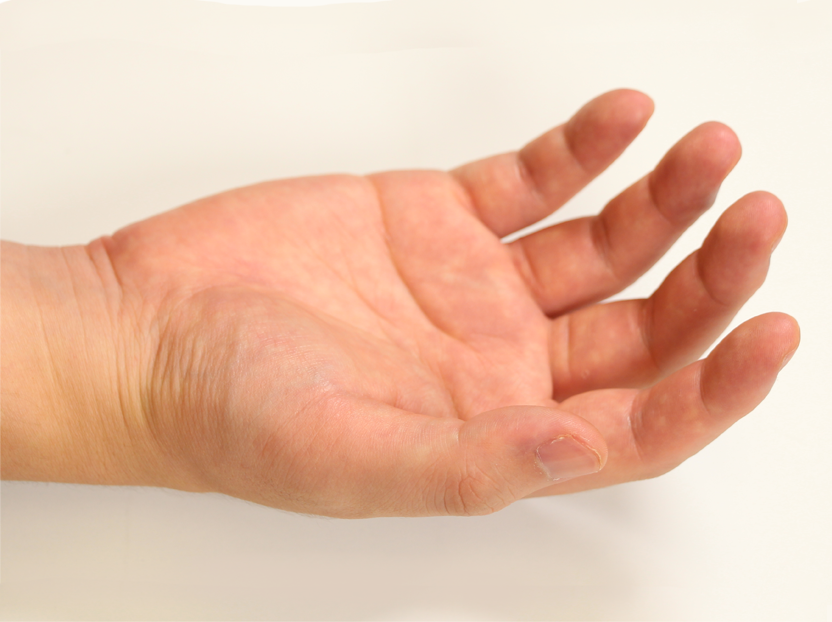 Pose 08 | 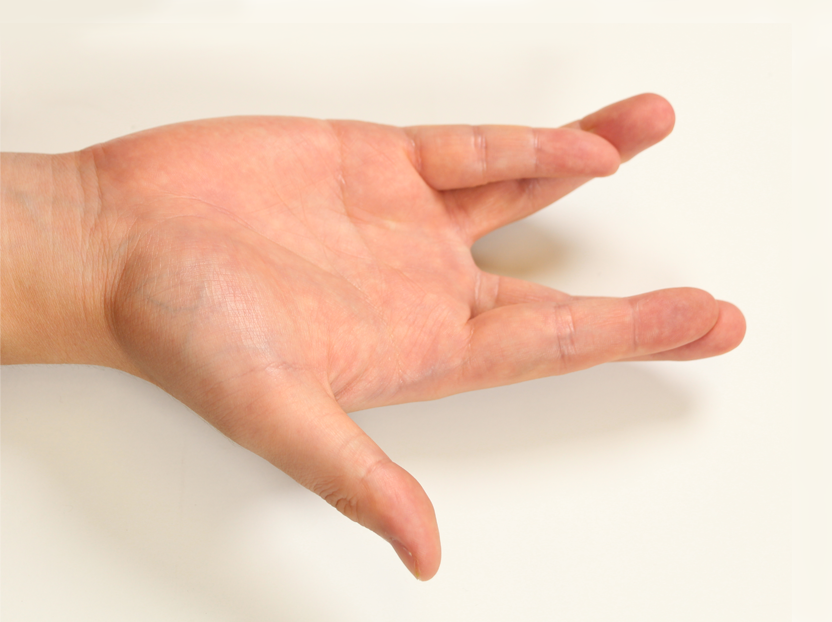 Pose 09 |  Pose 10 |  Pose 11 |  Pose 12 |
 Pose 01 |  Pose 02 |  Pose 03 |  Pose 04 |  Pose 05 |  Pose 06 |
 Pose 07 |  Pose 08 |  Pose 09 |  Pose 10 |  Pose 11 |  Pose 12 |
Where did you perform the MRI scans?
All MRI scans were performed on clinical MRI scanners in the Keck Medicine of USC hospital in Los Angeles, California, USA.
What are the specifications of your MRI machine and the MRI sequence used?
The images were obtained on a 3 Tesla GE MRI scanner using a PD CUBE (3D fast spin echo) sequence. Most scans of subject 1 were obtained in the sagittal plane; a few were obtained in the coronal plane. All scans of subjects 2, 3 and 4 were obtained in the coronal plane.
Parameters: NEX (number of excitations): 1 FA (flip angle): 90 degrees TR (time to repetition): 1500 ms TE (time to echo): 25.93 ms slice thickness: 1 mm slice spacing: 0.5 mm matrix: 256 x 256 pixels FOV (field of view): 25.6 x 25.6 cm
The scanner obtained 1 mm slice thickness samples in 0.5 mm overlapping slices. The obtained overlapping 1 x 1 x 1 mm voxels were automatically resampled to 0.5 x 0.5 x 0.5 mm isovoxel images (by the MRI machine software).
How did you select the 12 poses?
The 12 poses were selected to approximately cover the range of motion of the hand, and to actuate all important hand joints.
Who are the subjects?
The subjects are anonymous volunteers who consented to the scans and a public release of their data.
Subject 1: male, 28 years old. Height: 1.80m. Weight: 92kg. Subject 2: female, 48 years old. Height: 1.52m. Weight: 52kg. Subject 3: male, 45 years old. Height: 1.83m. Weight: 89kg. Subject 4: female, 26 years old. Height: 1.78m. Weight: 81kg.
What type of subjects did you recruit?
This dataset contains healthy hands free of musculoskeletal trauma or diseases. The female hand (Subject 02) has a minor localized swelling (about 1cm across) on the last link of the middle finger (pre-existing, temporary, and unrelated to our procedures and scans).
Why did you perform these scans?
To research human hand anatomy, to model how the anatomy (bones, muscles, fat, tendons, etc.) rigidly transforms and/or deforms as the hand actuates, to understand and model the function of the human hand, to animate and simulate human hands realistically.
Did you obtain IRB permission of your institution to release this dataset?
Yes. Prior to releasing this dataset, we obtained permission from our IRB, from all the subjects, our research sponsors, and the USC technology transfer office.
Can I use this dataset commercially?
Any use consistent with the license is permitted.
How much does it cost to use this dataset?
This dataset is free.
In what format is the data?
We give our MRI slices as PNG images. We use this format because it is extremely popular, and because it is both lossless and produces small files.
How can I get the exact intensity values from this dataset?
Our PNG images are single-channel 16-bit images and can be viewed in most popular image viewing programs (Photoshop, GIMP, IrfanView, etc.). In order to obtain the exact intensities, you need to write a computer program that opens the image using a PNG library (for example, libpng or stb). This will allow you to obtain the exact intensity values and use them in your application.
Why are the images in the downloaded dataset dark (for subjects 01 and 02) ?
For subjects 01 and 02, we used our own code to convert DICOM to PNG. Our MRI slices are single-channel 16-bit PNG images. Therefore, the intensity for each MRI pixel can in theory range from 0 to 65535. Our MRI machine produced integer intensity values in the range from 0 to approximately 3500. This does not fit into 8 bits, but fits into 16 bits. Using these values directly would result in very dark PNG images (when opened in an image viewing program) because 3500 is relatively small compared to the maximum possible 16-bit value of 65535. Therefore, we multiplied all intensities by 16 before exporting them to PNG images. We used the same value of 16 for all the MRI scans in this dataset. This covers the 0-65535 range better (but does not exceed it), and makes the images brighter. The images are still somewhat dark because the maximum intensity values occur rarely in our dataset. If you would like to make the images brighter, you can adjust the brightness in any image manipulation software.
For subjects 03 and 04, we used third-party software (OsiriX MD) to convert DICOM into PNG, and this resulted in brighter images.
Can you provide DICOM files?
DICOM files contain patient data, which we cannot provide for privacy reasons.